Common Loon
Common Loons (Gavia immer) are highly territorial, long-lived migratory species with limited breeding territory available. Common Loons exhibit specialized morphological traits that are uniquely adapted to stealthy diving; however, this makes terrestrial movement difficult. They occupy freshwater lakes in boreal and near-arctic habitats of North America and are sensitive to human disturbance, with disappearances from several former nesting areas due to human activities on lakes in summer. Moreover, Common Loons are projected to lose much of their breeding range as a result of climate change.
We teamed up with various partners (see below) to develop high-resolution molecular markers to identify breeding populations of Common Loons in North America. We scanned the genomes of 142 individuals from 23 sampling locations across the Common Loon breeding range to identify population structure. In total, we’ve identified the existence of 6 genetically distinct populations of the Common Loon – Alaska (green), Pacific Northwest (pink), central Canada (red), Midwest (yellow), eastern Canada (blue), and New England (purple). These 6 populations can serve as the foundation for Common Loon conservation and management.
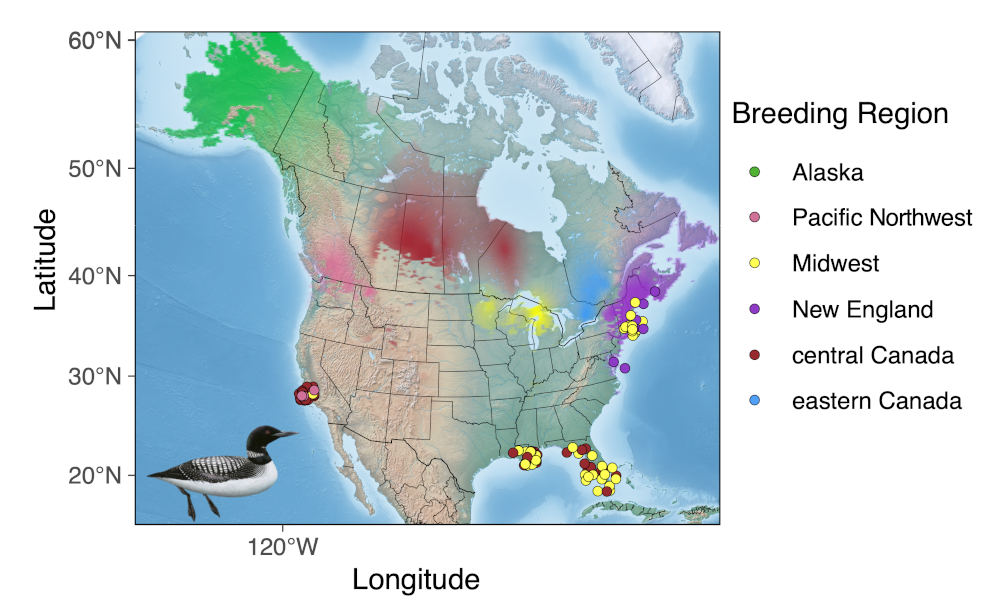
The six genetically distinct populations of Common Loon (Gavia immer) across the breeding range: Alaska (green), Pacific Northwest (pink), Midwest (yellow), New England (purple), Central Canada (red), and Eastern Canada (blue). Individuals sampled on the wintering grounds (colored dots) were then assigned back to their respective breeding ground genetic cluster, allowing us to better understand the migratory connectivity of Common Loon populations. Genetic clusters are visualized as transparency levels of different colors overlaid upon a base map from Natural Earth (naturalearthdata.com) and clipped to the species breeding range using an eBird shapefile. See below for details of samples used. Common Loon Image by © Birds of the World
Bird Migration Explorer: Click here to see how Audubon’s Bird Migration Explorer has incorporated our Common Loon genoscape into their base map.
Publications:
Larison B, Lindsay AR, Bossu C, Sorenson MD, Kaplan JD, Evers DC, Paruk J, DaCosta JM, and TB Smith. 2020. Leveraging genomics to understand threats in a migratory waterfowl. Evolutionary Applications 14: 1646-1658
Access to Sequencing Data:
Team:
Brenda Larson, Center for Tropical Research
Alec R. Lindsay, Northern Michigan University
Michael D. Sorenson, Boston University
Joseph D. Kaplan, Common Coast Research and Conservation
David C. Evers, Biodiversity Research Institute
James Paruk, Saint Joseph’s College
Jeffrey M. DaCosta, Boston College
R. Harrigan, UCLA
Utilized Common Loon Samples
Explore the map below to see when, where, and who collected the samples to build this genoscape.
