Wilson’s Warbler
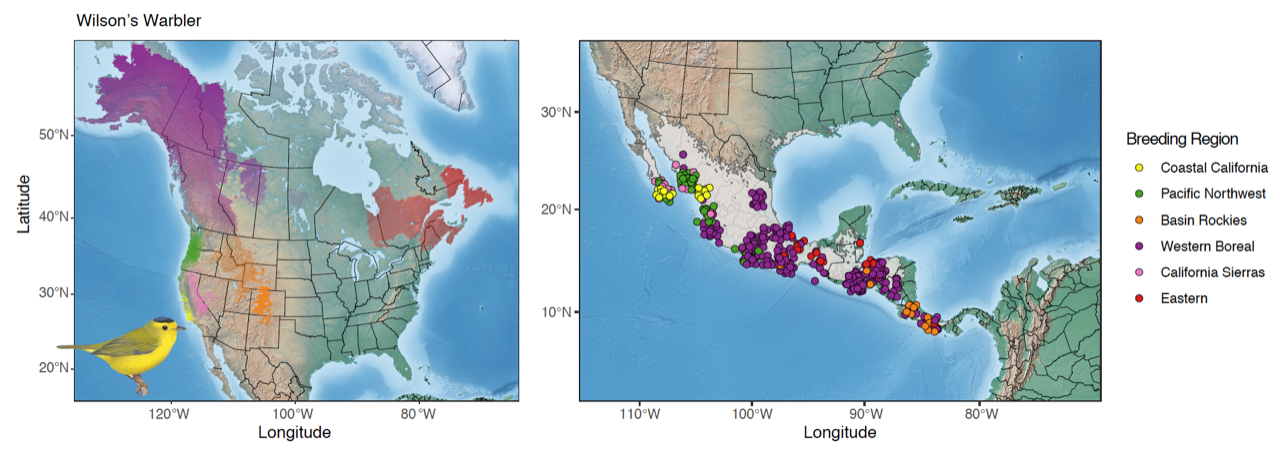
The six genetically distinct populations of Wilson’s Warbler (Cardellina pusilla) (left map) across the breeding range: Coastal California (yellow), Pacific Northwest (green), Basin Rockies (orange), Western Boreal (purple), California Sierras (pink), and Eastern (red). Individuals sampled on the wintering grounds (right map) were then assigned back to their respective breeding genetic cluster, allowing us to better understand the migratory connectivity of Wilson’s Warbler populations. Genetic clusters are visualized as transparency levels of different colors overlaid upon a base map from Natural Earth (naturalearthdata.com) and clipped to the species breeding range using an eBird shapefile. See below for details of samples used. Wilson’s Warbler Image by © Birds of the World
The Wilson’s Warbler (Cardellina pusilla) breeds in a wide range of habitats across Canada and the United States, but generally prefer alder and willow thickets, and forest edges with a dense understory. As these habitats disappear, population numbers have plummeted, resulting in a decline of over 60% since 1966. With their large breeding and wintering distributions, it has been complicated to understand which Wilson’s Warbler populations are facing the greatest declines.
The Bird Genoscape partnered with multiple collaborators (see below) to develop high-resolution molecular markers to identify breeding populations of Wilson’s Warblers in North America. We scanned the genomes of 408 individuals from 33 sampling locations across the Wilson’s Warbler breeding range to identify population genomic structure. These 6 populations can serve as the foundation for Wilson’s Warbler conservation and management.
Bird Migration Explorer: Click here to see how Audubon’s Bird Migration Explorer has incorporated our Wilson’s Warbler genoscape into their base map.
Publications:
Turbek SP, Polich A, Bossu CM, Rayne C, Carpenter A, Rodriguez Otero G, Gomez Villaverde S, Rodriguez Vasquez F, Hernandez-Banos BE, McCormack J, and Ruegg K. 2025. Genetic analysis of museum samples suggests temporal stability in the Mexican nonbreeding distribution of a neotropical migrant. Journal of Avian Biology, volume 1.
Ruegg KC, Harrigan RJ, Saracco JF, Smith TB, and CM Taylor. 2020. A genoscape-network model for conservation prioritization in a migratory bird. Conservation Biology 34: 1482-1491
Ruegg KC, Anderson EC, Harrigan RJ, Paxton KL, Kelly, Moore JF, F. and TB Smith. 2017. Genetic assignment with isotopes and habitat suitability (gaiah), a migratory bird case study. Methods in Ecology and Evolution: 8: 1241-1252
Ruegg KC, Anderson EC, Paxton KL, Apkenas V, Lao S, Siegel RB, DeSante DF, Moore F, and TB Smith. 2014. Mapping migration in a songbird using high-resolution genetic markers. Molecular Ecology 23: 5726-5739
Access to Sequencing Data:
https://github.com/eriqande/wiwa-popgen
https://datadryad.org/stash/dataset/doi:10.5061%2Fdryad.j5d33
Team:
Eric C. Anderson, Southwest Fisheries Science Center
Kristina L. Paxton, University of Hawaii, Hilo
Vanessa Apkenas, UCLA
Sirena Lao, Center for Tropical Research
Rodney B. Siegel, The Institute for Bird Populations
David F. DeSante, The Institute for Bird Populations
Frank Moore, University of Southern Mississippi
J.C. Garza
C.J. Ralph
L. West
D. Kaschube
P. Pyle
J. Saracco
R. Taylor
R. Fleischer
MAPs station operators within the Institute of Bird Populations Network
qb3 Genomics Sequencing Laboratory
Utilized Wilson’s Warbler Samples
Explore the map below to see when, where, and who collected the samples to build this genoscape.
